CRISPR used to build a biological hard drive out of live bacteria

Harvard scientists have found a way to store information on the DNA of living bacteria (Credit: destinacigdem/Depositphotos)
Hard drives will one day seem as primitive as punch cards or floppy disks, and it may turn out that the medium that replaces them was inside us all along: DNA. There's a mind-boggling amount of data naturally stored in the genome of every organism, with density and durability far beyond our best efforts. Now a Harvard team has created a biological hard drive, using the CRISPR gene-editing tool to record information to (and retrieve it from) the genome of living bacteria.
Scientists have been trying to tap into the storage potential of DNA for years. In 2012, Harvard genetics professor George Church encoded 70 billion copies of his book onto DNA, and later, researchers from Microsoft and the University of Washington broke a new record by storing 200 MB of data in DNA. Last year, Church and his team developed a molecular recorder based on the CRISPR system, which makes it easier to read and write information to and from the genome of bacteria.
Originally, the technology was designed to help scientists track the development of cells, by writing small but distinct changes to the genome. But creating a data storage device out of bacteria is a much larger undertaking, so the researchers set out to test how well their technique works to encode more complex information.
"As promising as this was, we did not know what would happen when we tried to track about 100 sequences at once, or if it would work at all," says Seth Shipman, first author of the study. "This was critical since we are aiming to use this system to record complex biological events as our ultimate goal."
While it's become synonymous with the gene-editing tool, CRISPR is the natural immune mechanism of bacteria. When a virus attacks a bacterium, samples of the viral DNA are captured and stored in the bacterium's genome as short "spacer" sequences, and the next time the bug encounters that strain of virus, this "memory" helps the immune protein Cas9 fight off the infection faster.
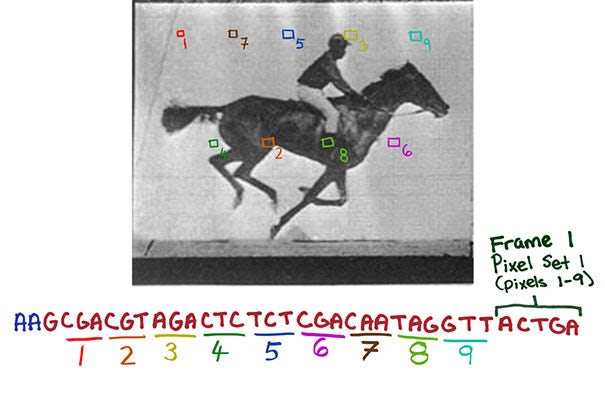
Individual pixels of each image were translated into snippets of DNA code, as well as the order in which each frame should appear, which allowed the digital data to be stored in the genome of living bacteria (Credit: Wyss Institute at Harvard University )
Since this process is effectively writing new information to the genome, the Harvard team found a way to hijack it, tricking the Cas1 and Cas2 proteins into storing specific information in these spacers. In this case, the data consisted of a digitized image of a human hand, and a short video of a galloping horse.
"We designed strategies that essentially translate the digital information contained in each pixel of an image or frame as well as the frame number into a DNA code, that, with additional sequences, is incorporated into spacers," says Shipman. "Each frame thus becomes a collection of spacers. We then provided spacer collections for consecutive frames chronologically to a population of bacteria which, using Cas1/Cas2 activity, added them to the CRISPR arrays in their genomes. And after retrieving all arrays again from the bacterial population by DNA sequencing, we finally were able to reconstruct all frames of the galloping horse movie and the order they appeared in."
The team plans to continue developing the technology to improve its use for data storage, as well as its original purpose of tracking cell changes over time.
The study was published in the journal Nature, and the researchers describe their work in the video below.
Source: Harvard
